Hosted by GEoPD member Dr. Manu Sharma and colleagues.
Tuebingen 2025
20th Annual Meeting of GEoPD | June 12–13, 2025
Hosted by GEoPD Member Dr. Manu Sharma
and colleagues.
Become a member of GEoPD
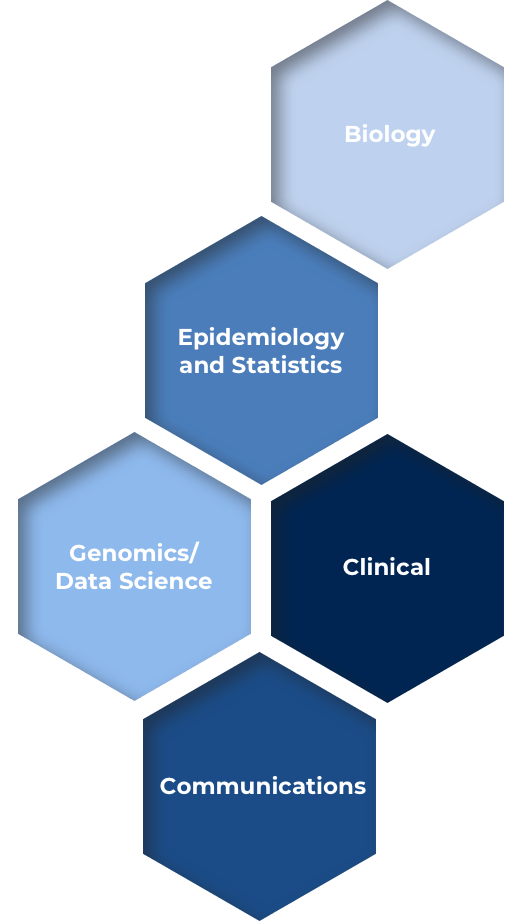
GEoPD is a global consortium of researchers dedicated to promoting education, scientific research and translational development in Parkinson’s disease. The consortium has been operating since 2004, and has an active membership from more than 60 sites on six continents. The GEoPD is organized into five cores.
These cores provide a mechanism whereby active global site investigators may share their expertise, resources and reagents. The consortium’s mission is to promote multi-investigator research projects throughout the year. Annual Meetings offer a valuable forum for consortium members to discuss unpublished data and ideas, highlight research questions or needs and identify global opportunities for partnership.
We truly appreciate your support in advancing PD research outreach and developing more effective treatment strategies.
Endorsement and Sponsors